Microbial diversity in the vaginal microbiota and its link to pregnancy outcomes

A prospective study of pregnant women
We established a prospective cohort of 2313 pregnant women with the goal of studying the vaginal microbiota, its community composition and the presence of pathogenic bacteria in the peripartum period. Inclusions and data collection were performed in accordance with applicable laws and ethical standards. Ethical approval for the study was obtained in November of 2017. Patients were enrolled, and samples collected, at three French hospitals in the Paris region (AP-HP): Hôpital Bichat Claude-Bernard, Hôpital Louis-Mourier and Hôpital Port-Royal. Informed consent was obtained prior to inclusion. In order to be included in the study, individuals had to have passed their 22nd week of gestation and be at least 18 years old. This work reports on 749 mothers whose pregnancies stared between 2018 and 2020, where vaginal swabs were collected through June of 2021 and whose clinical data records had been completed and validated.
The cohort, summarized in Fig. 1, was organized into four groups: (A) The control group of women who underwent standard-of-care screening at 32 weeks of pregnancy and who displayed no clinical risk of premature delivery or premature membrane rupture, (B) Full-term pregnancies in which there was a premature rupture of the membrane > 24 h before childbirth, (C) Premature births defined as labor before 37 weeks of gestation and (D) Premature deliveries that also experienced membrane rupture > 24 h before childbirth. Vaginal swabs were collected from the mother at the time of delivery.
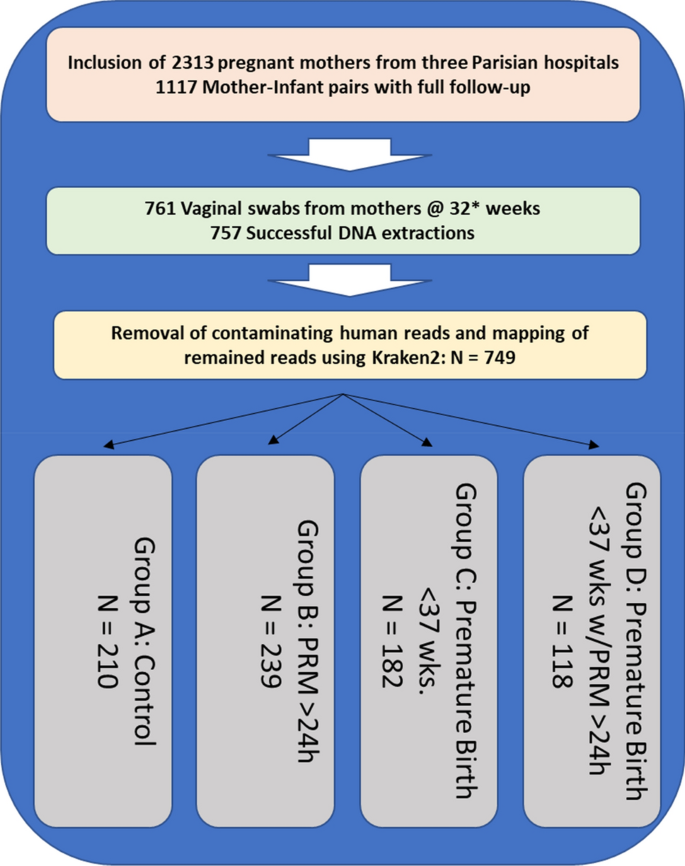
Cohort Composition and distribution of samples. 2313 pregnant mothers were recruited and consented to participate in the observational cohort. Current follow-up of mother-infant pairs with full clinical records equals 1117 completed dossiers. A total of 761 samples were selected for analysis. Four samples were excluded due to poor DNA extraction yield and a further eight samples did not yield sufficient sequence data to be included in the analysis. The distribution of samples across groups A, B, C and D is shown.
Categorical electronic patient records were used to collect both anthropomorphic as well as clinical data for individuals in the cohort, with the resulting information stored in a secure database. The health status of mothers was recorded using a set of variables used to identify complication or risk factors such as gestational diabetes, smoking or alcohol use during pregnancy, or high BMI. Bacteriological culture was performed, plating vaginal swabs on several media and allowing for the detection of Streptococcus agalactiae (group B Streptococcus, GBS) and additional potential pathogens including Escherichia coli, Klebsiella pneumonia, Proteus mirabilis, among others. Antibiotic susceptibility or resistance to a number of routine antibiotics such as macrolides (e.g., erythromycin) and beta-lactams (e.g., methicillin or carbapenem) were measured using both plating and PCR methods. Data collection continued after delivery for mother-infant pairs in order to track immediate or delayed pathogen infections across the four cohort arms. The initial focus of our analyses has been on the microbial composition of samples and links with risk to pregnancy outcomes and infection. A full listing of the analyzed samples in this study as well as the clinical variables used for this study are provided as supplemental data (Supplemental Table S1).
Dominant species influences pathogen presence in the vaginal microbiota
We analyzed maternal vaginal swabs from 761 of the completed mother-infant pairs. Sufficient DNA was successfully extracted from 757 samples. Samples were subsequently sequenced using Illumina paired ends (2 × 150 bp) technology. An average of 3.16 × 107 (SD \(\pm\) 1.4 × 107) high-quality reads were generated for each sample. Although bacteria play an important role in the vaginal environment, their biomass in relation to human cell is low. Consistent with this fact, we found that human host reads made up a mean of 95% of the total data set. Human-mapping reads were removed prior to performing downstream analysis. Taxonomic identification of filtered reads was determined using Kraken2 to map with our customized vaginal microbiota database. An average of 1.06 × 106 reads were mapped per sample. A total of 749 samples with matched patient records were retained for further analysis. Sequencing read identification and quantification for samples can be found in supplemental data (Supplemental Table S2).
We detected microbial communities of the vaginal microbiota mostly consistent with previously reported CSTs (community sequence types) (Fig. 2 and Supplemental Table S3). Lactobacillus crispatus (CST I) was the most abundant organism in our cohort, detected in 87% of samples and representing 30.18% of reads overall. Lactobacillus iners (CST III), 19.69% of total reads, was detected in 90.75% of samples and Gardnerella vaginalis (CST IV), 10.19% of total reads, was detected in 77.01% of samples. Lactobacillus jensenii (CST V), 3.42% of reads, and L. gasseri (CST II), 2.07% of reads, were also detected in > 75% of samples. Interestingly, we quantified several other bacterial species with relatively high total reads and high prevalence. Enterococcus faecalis was found in nearly 100% of samples and represented 3.88% of total reads. Pseudomonas tolaasii, 2.36% of reads, was detected in all samples while Escherichia coli, 2.36% of reads, was found in 83.75% samples. Streptococcus agalactiae (GBS), an important pathogen in both mothers and infants, was found in 74% of samples.
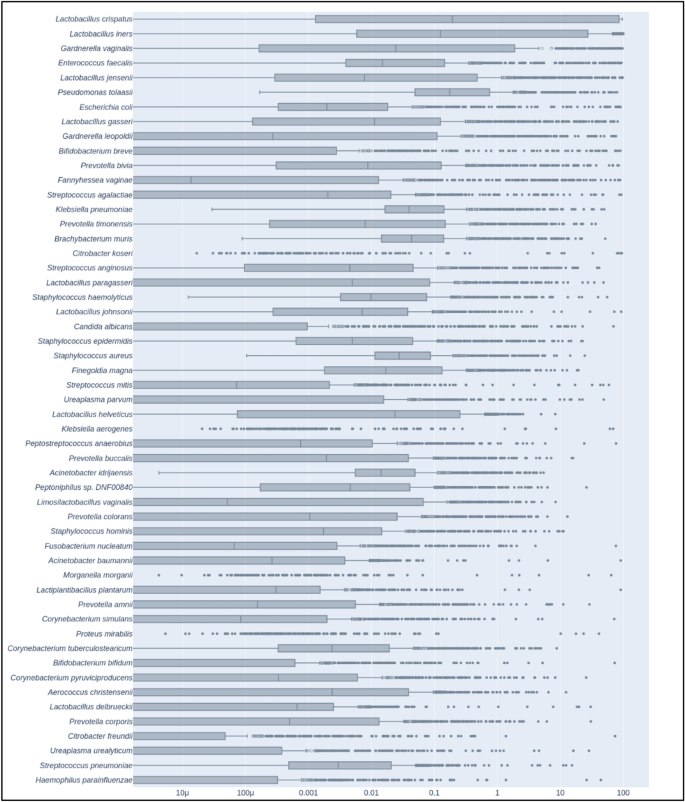
Relative abundance of the 53 most abundant microbial genomes among individuals of the cohort. Only species with a mean relative abundance across samples above 0.1% were retained for this graph. Boxes represent the interquartile range (IQR), delimited by the first and third quartiles (25th and 75th percentiles, respectively) and the line inside represents the median. Whiskers show the minimum and maximum values within 1.5 times IQR from the first and third quartiles respectively. Outliers (filled circles) are samples more than 3 times IQR below or above the first and third quartiles respectively. Suspected outliers (open circles) are samples between 1.5 and 3 times IQR below or above the first and third quartiles respectively.
Our examination of the most prevalent organisms revealed that, aside from the expected dominance of Lactobacillus species in normal vaginal flora, a large proportion of the remaining prevalent species detected were either known or opportunistic pathogens. This result was exemplified by the detection of Ureaplasma parvum in 45% of samples and U. urealyticum in 31% of samples. These two species have been previously observed in pregnant women and are associated with asymptomatic carriage as well as pregnancy complications including premature birth43. Candida albicans, another organism potentially associated with an elevated risk of preterm birth44, was found to be present in 41% of samples. Examining their co-occurrence, we found Candida and Ureaplasma genera are found together in 20.08% of the total cohort. We did not initially identify any significant differences in co-occurrence that could be directly linked with cohort groups for full-term versus pre-term delivery. However, an examination of clinical parameters revealed that when grouped by hospitalization reason, samples from medically programmed Cesarean deliveries showed a 26.67% co-occurrence rate compared with 17.84% for individuals admitted for spontaneous vaginal deliveries. Furthermore, when we investigated co-occurrence across CSTs, we found that L. jensenii, L. gasseri and L. crispatus communities had the lowest rate of co-occurrence at 9.52%, 11.76% and 15.14%, respectively. G. vaginalis communities displayed the greatest rate of co-occurrence at 34.62% followed by L. iners at 24.68%. A chi-squared test indicated the observed percentages to be significantly linked with CSTs (p = 4.04 × 10–4).
VMC Diversity and pregnancy outcomes
VMC diversity has been implicated in overall health and pregnancy outcomes with lower diversity community dominated by Lactobacillus species considered healthy with a lower risk of complications and infection45. We calculated the microbiota community α-diversity of vaginal samples using Faith’s phylogenetic diversity (PD) metric for the four groups of the cohort. Some differences were detectable in mean diversity between cohort groups (Control: PD = 20.2, PRM: PD = 19.0, Pre-term: PD = 22.1, Preterm with PRM: PD = 20.7), but these were not significant (Fig. 3a). A similar result was obtained when grouping samples according to hospitalization reason for mothers prior to delivery (Spontaneous Labor: PD = 20.4, Induced Labor: PD = 20.1, Membrane rupture: PD = 20.0, Risk of pre-term birth: PD = 22.3) (Fig. 3c). The elevated diversity in both the pre-term birth group as well as for the group hospitalized for an elevated risk of premature birth, while not significant, was nonetheless consistent with previous reports and was further investigated.
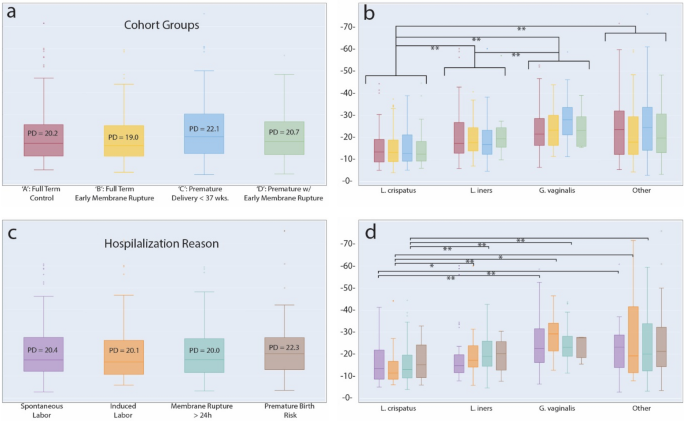
Faith’s Phylogenetic Diversity at the species level. (a) PD values for samples by ‘cohort group.’ No significant differences were found. (b) Samples grouped by dominant species, where a given species was most abundant in at least 100 samples. Each species group is subdivided by ‘cohort groups’ following the same color scheme as in ‘a’. Significant differences at the level of dominant species groupings are noted. (c) Samples grouped by ‘hospitalization reason’ for reasons with at least 40 samples. No significant differences were found. (d) Samples grouped by dominant species and subdivided by ‘hospitalization reason’ following the same color scheme as in ‘c’. *p < 0.05; **p < 0.01; False Discovery Rate, correction with Benjamini–Hochberg.
Having detected significant differences in VMC structure across CSTs, we followed up and stratified samples by their dominant bacterial species: L. crispatus, L. iners, and G. vaginalis. A fourth group, ‘other,’ included samples not dominated by one of these species. By using these bacterial groups, significant differences in diversity were uncovered, and were most striking, when comparing L. crispatus communities to the other three groupings (L. crispatus PD = 15.4, L. iners PD = 20.1, G. vaginalis PD = 25.1, ‘other’ PD = 23.7) (Fig. 3b). Independent Wilcoxon tests indicated significant differences between L. crispatus and L. iners samples (p = 7.02 × 10–8) and L. crispatus and G. vaginalis samples (p = 5.75 × 10–17). A weaker, but still significant difference existed when comparing L. iners and G. vaginalis (p = 9.72 × 10–6). Interestingly, there was no significant effect on diversity result when ‘other’ community types were calculated as either exclusive or inclusive of Lactobacilli from other common CSTs: L. gasseri (CST II) and L. jensenii (CST V). We also detected a number of significant differences in diversity associated with specific hospitalization reason, again primary in relation to samples with a L. crispatus dominance (Fig. 3d). A full accounting of comparisons and significant differences can be found in Supplemental Table S5.
Further examination of the bacterial composition of samples yielded significant differences in the distribution of mothers according to hospitalization reason. We found that individuals admitted for pre-term delivery risk had a VMC dominated by taxa ‘other’ than L. crispatus, L. iners, or G. vaginalis, compared with admission full-term spontaneous deliveries (chi-squared, p = 7.87 × 10–4). Interestingly, we did not identify a significant difference specifically linked to G. vaginalis and hospitalization reason. Given the significant association with a VMC that appeared potentially dysbiotic, outside of common CSTs, we next looked at reported pregnancy outcomes. Chi-squared testing of samples, binned this time by pregnancy length, showed that a significantly (p = 1.02 × 10–4) higher number of samples with shorter pregnancy lengths had VMCs dominated by bacterial species other than L. crispatus, L. iners, or G. vaginalis (Supplemental Figure S1). The p-value decreased to 2.84 × 10–8 when excluding all other Lactobacillus-dominated samples. These results, summarized also in Supplemental Table S6, suggest that a combination of non-standard bacterial community combined with a higher overall diversity could be markers of an increased risk of premature delivery.
Dominant taxa are associated with clinical outcomes
In order to further define the role of community types and diversity, we performed PCoA analysis46 using the normalized taxonomic frequency data from vaginal microbiota samples. Bray–Curtis dissimilarity distances were generated that included all samples and this data projected in two-dimensions. The resulting structure of the PCoA corroborated our observation of VMCs dominated by L. crispatus, L. iners and G. vaginalis (Fig. 4a). We observed that 20.15% of sample variability could be explained by on the primary axis (PC1). The top taxa loading for each axis was plotted as vectors; For PC1, the transition between the two dominant Lactobacillus species appeared to be the most important factor separating the samples. Generally, samples could be distinguished by the community abundance of L. crispatus versus non-L. crispatus species (moving left to right). Figure 4b represents the same data with coordinates plotted for the second and third principal coordinates (PC2 & PC3). Here, two other dominant communities, Gardnerella vaginalis and L. iners, are clearly separated across the top of the plot and a third community with a strong loading for E. coli is placed in the lower half. Interestingly, Klebsiella, a genus of opportunistic human pathogens, was observed with a loading similar to E. coli, and distanced from the standard CST species. This again suggested that a certain amount of opportunistic pathogen diversity could be associated with divergence from the canonical community types.
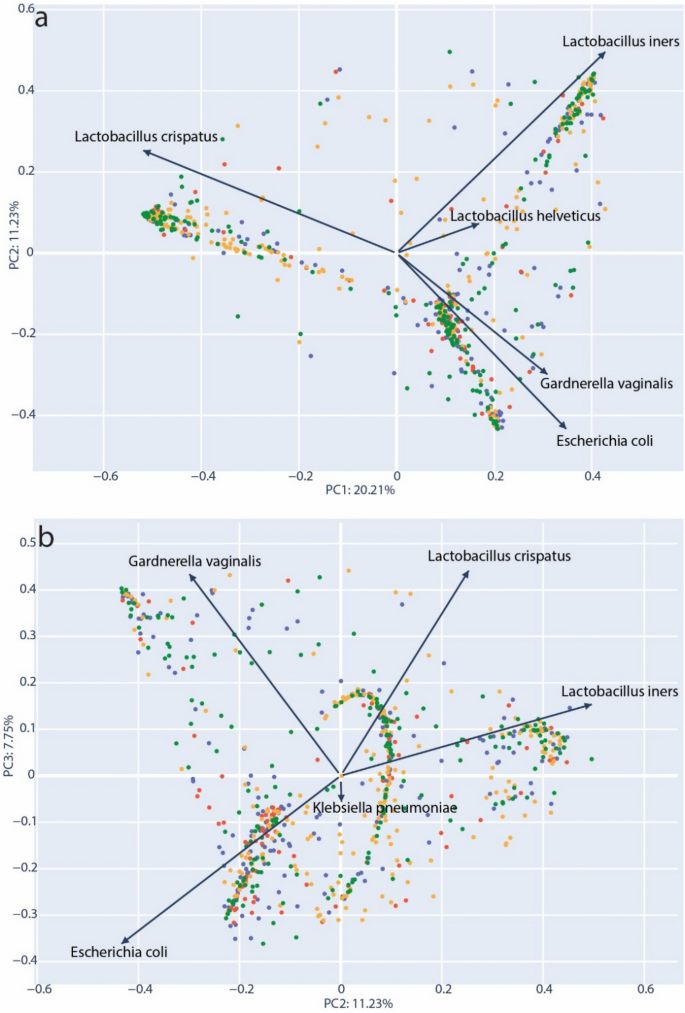
PCoA projection of Bray–Curtis dissimilarity. (a) Principal components PC1 and PC2 used for 2D projection for samples. (b) PC2 and PC3 used for 2D projection of samples. Individual components (species) that are the primary drivers along each axis are indicated with an arrow representing the strength of each loading proportional to its length. Samples are colored according to their inclusion group: green = control full-term delivery, orange = membrane rupture > 24 h prior to full-term delivery, blue = preterm delivery < 37 weeks, red = preterm delivery with membrane rupture > 24 h prior to delivery.
We performed PERMANOVA analysis to identify significant groupings based on clinical data for samples in the Bray–Curtis dissimilarity matrix. An analysis of all samples (n = 749) at the species level showed that only the abundance of Haemophilus influenzae was significantly associated with the Bray–Curtis groupings (p = 0.038). However, when the analysis was performed by first stratifying samples by their most abundant taxa, we found other significant associations. L. iners-dominated samples (n = 160) were found to have significant grouping for both hospitalization reason (p = 0.008) and birth mode (p = 0.043). Within G. vaginalis-dominated samples (n = 104), analyzed at the genus level, there were significant associations for frequent urinary tract infections (p = 0.044), umbilical cord inflammation (p = 0.042) and gestation length at time of admittance (p = 0.022). It is notable that L. crispatus-dominated samples (n = 251) yielded no significant clinical associations.
Random forest classifier
Identification of links between microbial diversity and pregnancy length coupled to associations of dominant VMC species to clinical variables of risk, including gestation time and infection, were extremely interesting. We used machine learning as an efficient means to further explore these results through selection and integration of a large number of microbial variables. A Random Forest Classifier (RFC) was trained to integrate the contributions of multiple members of the VCM in order to explain clinical observations. We focused on hospitalization for a risk of premature birth, an important variable in the study.
There was a total of 48 samples from mothers admitted to the hospital due to an elevated risk of premature birth. An equal number of control samples was randomly selected from mothers admitted under the standard procedure for spontaneous labor (Supplemental Table S4). We trained the RFC model on a subset of 133 species variables that were present in at least 25% of samples and which also had a minimum of 50 mean normalized reads across all samples. A preliminary round of training resulted in a classifier that incorporated the frequencies of 109 species variables. The species feature with the highest importance in the 109-species model was Mageeibacillus indolicus, a recently isolated bacterium from the female genital tract47. Recursive factor removal (RFR) was performed whereby the classifier was refined by eliminating the least important species variable from the data set and retraining with the remaining taxa. This process was run iteratively, testing the accuracy of the resulting classifier at each round. Local regression (LOWESS) applied to a scatterplot of model accuracy indicated that a maximal accuracy was achieved with twelve species variables (Supplemental Table S4). These twelve species were then used to train the definitive classifier. The overall accuracy of the RFC in distinguishing admission for full-term spontaneous births from samples admitted for a risk of premature birth increased from 62.5% percent for the preliminary model with 109 variables to 83.3% with the optimized twelve species. For validation, a receiver operating characteristic (ROC) curve was constructed from three independent combinations of training and testing samples. ROC analysis yielded a mean area under the curve (AUC) value of 0.82 ± 0.04 (Fig. 5a). The twelve most informative species variables (Fig. 5b) are, excepting Peptoniphilus lacrimalis, gram-positive with the top three (Streptococcus mitis, Streptococcus agalactiae, Fusobacterium nucleatum) known as potential pathogens. Notably, the Lactobacillus species in the list (L. delbrueckii, L. paragasseri, L. reuteri, L. anylolyticus) are not those which normally predominate the major CSTs.
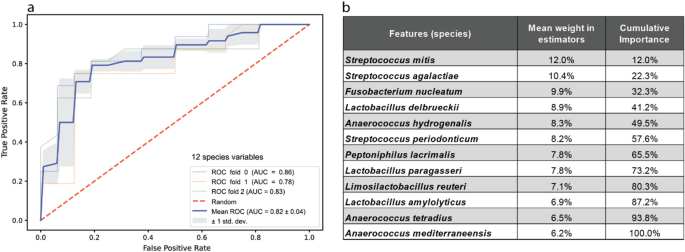
Random Forest predictor of premature birth risk. (a) Receiver operator characteristic (ROC) test of the RF classifier for normal and high-risk hospital admissions. Samples are class scored as correctly classified ‘true positives’ or incorrectly predicted ‘false positives.’ Areas under the curve are calculated for three random mixtures of training and test samples, blue, orange, and green lines, compared to an expected random result, dashed red line. (b) List of the twelve species components in the best performing RF classifier. The mean weights over individual estimators are given as well as their cumulative weighs in the final predictor.












